import os
import mne
import numpy as np
import matplotlib.pyplot as plt
import pickle
import pandas as pd
from scipy import signal
from scipy.io import wavfile
# from pybv import write_brainvision
from pyprep.prep_pipeline import PrepPipeline
from mne_icalabel import label_componentsCodeswtich preprocessing pipeline
1 Libraries
2 Trigger lag fix
2.1 parameters
# directory
input_dir = os.getcwd() + '/data/'
output_dir = os.getcwd() + '/preprocessed/1_trigger_lag_corrected/'
# create a folder if the folder doesn't exist
os.makedirs(output_dir, exist_ok=True)
# subjects to exclude
exclude_subs = [
]
# trigger searching window (actual trigger time based on audio - trigger time in the data)
t_left = -0.01
t_right = 0.5Here we fix trigger lag fix and recode the event for item-level analysis.
#### create dictionaries for item-level codes and descriptions ####
df = pd.read_csv("mapping_file.txt", delimiter='\t')
mapping_file2code = dict(zip(df['filename'], df['item_code']))
mapping_code2description = dict(zip(df['item_code'], df['description']))
#############################################################
# get list of file names
all_files = os.listdir(input_dir)
# for each file
for file in all_files:
if file.endswith('.vhdr') and (file.split('.')[0] not in exclude_subs) and (file.split('.')[0]+ '_corr.fif' not in os.listdir(output_dir)):
# read in vhdr files
raw = mne.io.read_raw_brainvision(input_dir + file, preload = True)
# extract sampling rate
eeg_sfreq = raw.info['sfreq']
#### get trigger code, audio data, and audio length ####
# initialize dictionaries
trigger_dict = {} # marker: [description list]
audio = {} # filename: data
lengths = {} # filename: audio length
# read in the mapping file
with open('mapping_file.txt','r') as f:
# skip the first line (header)
next(f)
for line in f:
# read in the current line
line = line.replace('\n','')
# get info
filename, marker, description, item_code = line.split('\t')
# initialize a filename list for each trigger code
if marker not in trigger_dict.keys():
trigger_dict[marker] = []
# add the filename to the list
trigger_dict[marker].append(filename)
# get audio data for each file
if filename not in audio.keys():
# get sample rate and data of the audio file
sampleRate, data = wavfile.read('codeswitch_mystim/stimuli/{}'.format(filename))
# the sound file is stereo, so take only 1 channel's data
data = data[:,0]
# calculate sound file length
lengths[filename] = len(data)/sampleRate
# reduce the sampling rate of the audio file by the factor of int(sampleRate/eeg_sfreq)
data_downsampled = signal.decimate(data, int(sampleRate/eeg_sfreq), ftype='fir')
# add audio data the audio dictionary
audio[filename] = data_downsampled
####################################################
#### get events ####
# for each stimulus, mark the block info
events_from_annot, event_dict = mne.events_from_annotations(raw, verbose='WARNING')
# only events with trigger code 1-4 are useful
events_from_annot = events_from_annot[events_from_annot[:, 2] <= 4]
######################
#### cross correction to find the audio file and correct lag correction ####
# initialize
delays = np.array([]) # a delay list
bad_stim = [] # a bad stim list
corr_results = [] # list of each event's max cross-correlation coefficient
filename_results = [] # list of each event's filename
# loop over each event
for i in range(len(events_from_annot)):
# get current event
event = events_from_annot[i]
# get the onset latency
time = event[0]/eeg_sfreq
# get the marker
marker = str(event[2])
# initialize dictonary of each file and its max correlation coefficent
singleFile_maxCorr_dict = {}
# initialize dictionary for each file and its lag corresponding to the max correlation coefficent
singleFile_maxCorrLag_dict = {}
#### find the audio file for the current event, recode the marker, and record the lag info ####
for name in trigger_dict[marker]:
# get the data from the sound channel
audio_eeg = raw.get_data(
picks = ['StimTrak'],
tmin = time + t_left,
tmax = time + lengths[name] + t_right,
)[0]
# get actual stimulus data
audio_stim = audio[name]
# z-score normalization (subtract mean, divide by std)
audio_eeg = (audio_eeg - np.mean(audio_eeg)) / np.std(audio_eeg)
audio_stim = (audio_stim - np.mean(audio_stim)) / np.std(audio_stim)
# cross-correlation
corr = signal.correlate(audio_eeg, audio_stim, mode='full')
# normalize for signal duration
corr = corr / (np.linalg.norm(audio_eeg) * np.linalg.norm(audio_stim))
# find peak correlation value
singleFile_maxCorr_dict[name] = np.max(corr)
# get lags for cross-correlation
lags = signal.correlation_lags(
audio_eeg.size,
audio_stim.size,
mode="full")
# find the lag for peak correlation
singleFile_maxCorrLag_dict[name] = lags[np.argmax(corr)] + t_left*eeg_sfreq
# get the file giving max correlation
max_file = max(singleFile_maxCorr_dict, key=singleFile_maxCorr_dict.get)
# get the maximum correlation among all files
max_corr = singleFile_maxCorr_dict[max_file]
# get the lag
lag = singleFile_maxCorrLag_dict[max_file]
# add item-level trigger code
events_from_annot[i][2] = mapping_file2code[max_file]
# if the maximum correction is less than a threshold
if round(max_corr,1) < 0.5:
# mark the stim bad
bad_stim.append(i)
# add the maximum correlation info for the current event
corr_results.append(max_corr)
filename_results.append(max_file)
delays = np.append(delays,lag)
##################################################
#### plot the stimtrak eeg and the audio data of the event with the minimum correlation of the current file ####
# get min corr
min_corr = np.argmin(corr_results)
# get current event info
event = events_from_annot[min_corr]
# get the onset latency
time = event[0]/eeg_sfreq
# get the file name of the event
name = filename_results[min_corr]
# get the stimtrak data
audio_eeg = raw.get_data(
picks = ['StimTrak'],
tmin = time + t_left,
tmax = time + lengths[name] + t_right,
)[0]
# actual stimulus data
audio_stim = audio[name]
# z-score normalization (subtract mean, divide by std)
audio_eeg = (audio_eeg - np.mean(audio_eeg)) / np.std(audio_eeg)
audio_stim = (audio_stim - np.mean(audio_stim)) / np.std(audio_stim)
# plot
fig, ax = plt.subplots()
ax.plot(audio_eeg, label = 'StimTrak', alpha = 0.6)
ax.plot(audio_stim, label = 'wave', alpha = 0.6)
ax.set_title(file)
ax.legend()
fig.savefig(output_dir + file.split('.')[0] + "_minCor.png", dpi=300, bbox_inches='tight')
##########################
# record number of bad stims of the current file
if len(bad_stim)>0:
# wave the number of bad stims to a file
with open(output_dir + 'bad_stim.txt', 'a+') as f:
_ =f.write(file + '\t' + str(len(bad_stim)) + ' bad stims' + '\n')
# remove events of bad stims
events_from_annot = np.delete(events_from_annot, bad_stim, 0)
# remove lags of bad stims
delays = np.delete(delays, bad_stim, 0)
# correct for trigger lag
events_from_annot[:,0] = events_from_annot[:,0] + delays
# create item-level annotations
annot_from_events = mne.annotations_from_events(
events = events_from_annot,
event_desc = mapping_code2description, # item-level mapping
sfreq = eeg_sfreq
)
# set annotations
raw.set_annotations(annot_from_events)
# drop the audio channel in data
raw.drop_channels(['StimTrak'])
# save single-trial delay file
np.savetxt(output_dir + file.replace('.vhdr', '_delays.txt'), delays, fmt='%i')
# save as a file-into-file data
raw.save(output_dir + file.split('.')[0]+ '_corr.fif')3 Bad channel correction
- filtering
- resampling
- remove line noise
- bad channel detection & repairing
- add back reference channel TP9
3.1 parameters
#### parameters ####
# set directory
input_dir = os.getcwd() + '/preprocessed/1_trigger_lag_corrected/'
output_dir = os.getcwd() + '/preprocessed/2_bad_channel_corrected/'
# create a folder if the folder doesn't exist
os.makedirs(output_dir, exist_ok=True)
# filter cutoff frequencies (low/high)
f_low = 1
f_high = 100
# resampling frequency
f_res = 250
# line frequency
line_freq = 60
# preprocessing parameters
prep_params = {
"ref_chs": 'eeg',
"reref_chs": 'eeg', # average re-reference
"line_freqs": np.arange(line_freq, f_res/2, line_freq),
}
# create a montage file for the pipeline
montage = mne.channels.make_standard_montage("standard_1020")
# interpolation method
# method=dict(eeg="spline")#####################################################
#### Preprocessing (filtering, resampling, bad channel detection/interpoloation, re-reference) ####
#####################################################
# get all file namesin the folder
all_input = os.listdir(input_dir)
all_output = os.listdir(output_dir)
# for each file
for file in all_input:
if file.endswith("corr.fif") and (file.split('.')[0]+ '_prep.fif' not in all_output):
# read in file
raw = mne.io.read_raw_fif(input_dir + file, preload=True)
# set channel type for EOG channels
raw.set_channel_types({'Fp1':'eog', 'Fp2':'eog'})
# filter
raw.filter(l_freq = f_low, h_freq = f_high)
#### cut off the beginning and ending part ####b
# get the onset of the first and the last event ####
events_from_annot, event_dict = mne.events_from_annotations(raw, verbose='WARNING')
#### crop the file to cut off the first the last 10s portion which maybe noisy ####
# define the beginning time (in seconds)
crop_start = events_from_annot[0][0]/raw.info['sfreq'] - 10
# define the ending time (in seconds)
crop_end = events_from_annot[-1][0]/raw.info['sfreq'] + 10
# crop the data
raw.crop(
tmin=max(crop_start, raw.times[0]),
tmax=min(crop_end, raw.times[-1])
)
####################################################################################
# resample
raw.resample(sfreq = f_res)
# read in channel location info
raw.set_montage(montage)
#### Use PrePipeline to preprocess ####
'''
1. detect and interpolate bad channels
2. remove line noise
3. re-reference
'''
# apply pyprep
prep = PrepPipeline(raw, prep_params, montage, random_state=42)
prep.fit()
# export a txt file for the interpolated channel info
with open(output_dir + 'bad_channel.txt', 'a+') as f:
_ =f.write(
file + ':\n' +
"- Bad channels original: {}".format(prep.noisy_channels_original["bad_all"]) + '\n' +
"- Bad channels after robust average reference: {}".format(prep.interpolated_channels) + '\n' +
"- Bad channels after interpolation: {}".format(prep.still_noisy_channels) + '\n'
)
# save the pypred preprocessed data into the raw data structure
raw = prep.raw
# add back the reference channel
raw = mne.add_reference_channels(raw,'TP9')
# add the channel loc info (for the newly added reference channel)
raw.set_montage(montage)
# save
raw.save(output_dir + file.split('.')[0]+ '_prep.fif')4 ICA artifact subtraction
4.1 parameters
# directory
input_dir = os.getcwd() + '/preprocessed/2_bad_channel_corrected/'
output_dir = os.getcwd() + '/preprocessed/3_ica/'
# create a folder if the folder doesn't exist
os.makedirs(output_dir, exist_ok=True)
# up to which IC you want to consider
ic_upto = 15
# ic_upto = 99# get all file names in the folder
all_input = os.listdir(input_dir)
all_output = os.listdir(output_dir)
# initialize a dictionary for files
for file in all_input:
if file.endswith("prep.fif") and (file.split('.')[0] + '_ica.fif' not in all_output):
# read in file
raw = mne.io.read_raw_fif(input_dir + file, preload=True)
# make a filtered file copy ICA. It works better on signals with 1 Hz high-pass filtered and 100 Hz low-pass filtered
raw_filt = raw.copy().filter(l_freq = 1, h_freq = 100)
# apply a common average referencing, to comply with the ICLabel requirements
raw_filt.set_eeg_reference("average")
# initialize ica parameters
ica = mne.preprocessing.ICA(
# n_components=0.999999,
max_iter='auto', # n-1
# use ‘extended infomax’ method for fitting the ICA, to comply with the ICLabel requirements
method = 'infomax',
fit_params = dict(extended=True),
random_state = 42,
)
#### get ica solution ####
ica.fit(raw_filt, picks = ['eeg'])
# save ica solutions
ica.save(output_dir + file.split('.')[0]+ '_icaSolution.fif')
#### ICLabel ####
ic_labels = label_components(raw_filt, ica, method="iclabel")
# save
with open(output_dir + file.split('.')[0]+ '_icLabels.pickle', 'wb') as f:
pickle.dump(ic_labels, f)
#### auto select brain AND other ####
labels = ic_labels["labels"]
exclude_idx = [
idx for idx, label in enumerate(labels) if idx<ic_upto and label not in ["brain", "other"]
]
# ica.apply() changes the Raw object in-place
ica.apply(raw, exclude=exclude_idx)
# record the bad ICs in bad_ICs.txt
with open(output_dir + '/bad_ICs.txt', 'a+') as f:
_ = f.write(file + '\t' + str(exclude_idx) + '\n')
# save data after ICA
raw.save(output_dir + file.split('.')[0]+ '_ica.fif')5 Segmentation
segmenting continuous EEG into epochs - re-reference - segmentation
5.1 parameters
#### parameters ####
# directory
input_dir = os.getcwd() + '/preprocessed/3_ica/'
output_dir = os.getcwd() + '/preprocessed/4_erp_epochs/' # for ERP
# create a folder if the folder doesn't exist
os.makedirs(output_dir, exist_ok=True)
# reject data
reject_subs = [
]
# Epoch window:
erp_t_start = -0.2; erp_t_end = 0.8
baseline = (-0.2, 0)
# criteria to reject epoch
# reject_criteria = dict(eeg = 100e-6) # 100 µV
# reject_criteria = dict(eeg = 150e-6) # 150 µV
reject_criteria = dict(eeg=200e-6) # 200 µV# epochs for ERP
# initialize a list for subjects with too many bad trials
too_many_bad_trial_subjects = []
# get file names
all_input = os.listdir(input_dir)
all_output = os.listdir(output_dir)
#### re-reference, and epoch ####
for file in all_input:
if file.endswith("ica.fif") and (file.split('.')[0] + '_epo.fif' not in all_output):
# skip the rejected subject
if file.split('_')[1] in reject_subs:
continue
# read in data
raw = mne.io.read_raw_fif(input_dir + file, preload = True)
# average-mastoids re-reference
raw.set_eeg_reference(ref_channels = ['TP9', 'TP10'])
#### this is for source calculation ####
# filter the data, optional
# raw = raw.filter(l_freq=None, h_freq=30)
# sphere = mne.make_sphere_model('auto', 'auto', raw.info)
# src = mne.setup_volume_source_space(sphere=sphere, exclude=30., pos=15.)
# forward = mne.make_forward_solution(raw.info, trans=None, src=src, bem=sphere)
# raw = raw.set_eeg_reference('REST', forward=forward)
########################################
# get event info for segmentation
events_from_annot, event_dict = mne.events_from_annotations(raw, verbose='WARNING')
# segmentation for ERP
epochs = mne.Epochs(
raw,
events = events_from_annot, event_id = event_dict,
tmin = erp_t_start, tmax = erp_t_end,
# apply baseline correction
baseline = baseline,
# remove epochs that meet the rejection criteria
reject = reject_criteria,
preload = True,
)
# for each event, remove 0 trial events, record info, and check if a subject is bad
for k, v in event_dict.items():
# good trial count
trial_count = len(epochs[k])
# remove 0 trial event
if trial_count==0:
del epochs.event_id[k]
# good trial rate
goodTrial_rate = round( trial_count/sum(events_from_annot[:,2]==v), 2 )
# record epoch summary
with open(output_dir + 'epoch_summary.txt', 'a+') as f:
_ =f.write(file.split('_')[1] + '\t' + k + '\t' + str(trial_count) + '\t' + str(goodTrial_rate) + '\n')
# mark a subject bad if any condition has fewer than 1/2 trials
if ( goodTrial_rate < 0.5 ):
# mark the subject file as bad
if file.split('_')[1] not in too_many_bad_trial_subjects:
too_many_bad_trial_subjects.append(file.split('_')[1])
# save single subject file
epochs.save(output_dir + file.split('.')[0] + '_epo.fif',
overwrite=True)
# export the record of bad subjects for ERP
with open(output_dir + 'too_many_bad_trial_subjects.txt', 'w') as file:
# Write each item in the list to the file
for item in too_many_bad_trial_subjects:
file.write(item + '\n')6 ERP
6.1 parameters
# directory
input_dir = os.getcwd() + '/preprocessed/4_erp_epochs/'
output_dir = os.getcwd() + '/preprocessed/5_averaged/'
# create a folder if the folder doesn't exist
os.makedirs(output_dir, exist_ok=True)#### get ERP ####
# get file names
all_input = os.listdir(input_dir)
all_output = os.listdir(output_dir)
# initialize a dictionary to store data
all_evokeds = {}
# bad subjects with 0 good trials in any condition
bad_subs = [
]
# for each file
for file in all_input:
if file.endswith("_epo.fif"):
# extract subject number
subject = file.split('_')[1]
# skip rejected subjects
if subject in bad_subs:
continue
# read in data
epochs = mne.read_epochs(input_dir + file, preload = True)
# average | get ERP for each condition
evoked = epochs.average(by_event_type=True)
# initialize dictionary for single-subject ERP
all_evokeds[file.split('_')[1]] = {}
# add key for each condition for analysis
for cond in evoked:
# append the evoked data to the dictioncary of evoked data
all_evokeds[file.split('_')[1]][cond.comment] = cond
# Saving the ERP data:
with open(output_dir + '/all_evokeds.pkl', 'wb') as f:
pickle.dump(all_evokeds, f)
# del all_evokeds7 Visulization
7.1 parameters
# directory
input_dir = os.getcwd() + '/preprocessed/5_averaged/'
# participants to exclude
exclude_ppts = []7.2 single-participant, single-condition butterfly
# read in the ERP data:
with open(input_dir + '/all_evokeds.pkl', 'rb') as file: # Python 3: open(..., 'rb')
all_evokeds = pickle.load(file)
# get data
evoked = all_evokeds['836']['local_noswitch_lunchbox']
# waveform
evoked.plot()
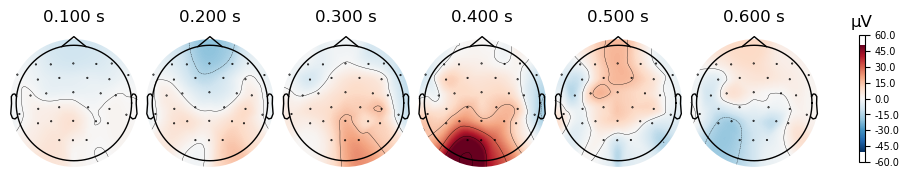
# scalp topography
times = [0.1, 0.2, 0.3, 0.4, 0.5, 0.6]
evoked.plot_topomap(times=times, colorbar=True)
plt.show()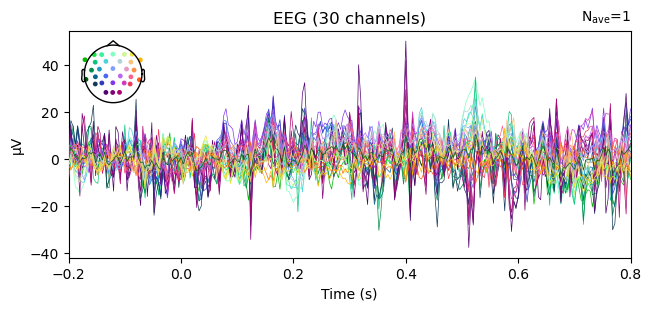
7.3 topographical subplots
# read in the ERP data:
with open(input_dir + '/all_evokeds.pkl', 'rb') as file:
all_evokeds = pickle.load(file)
# get the list of all participants that came this far
all_ppts = list(all_evokeds.keys())
# get participants that meet criteria
sub_ppts = []
for ppt in all_ppts:
# if it is not in the bad subject list #
if ppt not in exclude_ppts:
# append that subject to the list
sub_ppts.append(ppt)
# extract ERPs for each condition
local_switched = []
local_noswitch = []
mando_switched = []
mando_noswitch = []
# for each participant
for ppt in sub_ppts:
# extract item labels
items = all_evokeds[ppt].keys()
for cond in ['local_switched', 'local_noswitch', 'mando_switched', 'mando_noswitch']:
# get condition list
cond_list = [ x for x in items if x.rsplit('_', 1)[0]==cond ]
# compute erp
tmp= mne.combine_evoked([all_evokeds[ppt][x] for x in cond_list],
weights='equal')
# append erp to list
eval(cond).append(tmp)
# add erp data to dictionary for plotting
evokeds = {}
for cond in ['local_switched', 'local_noswitch', 'mando_switched', 'mando_noswitch']:
evokeds[cond] = eval(cond)
################################
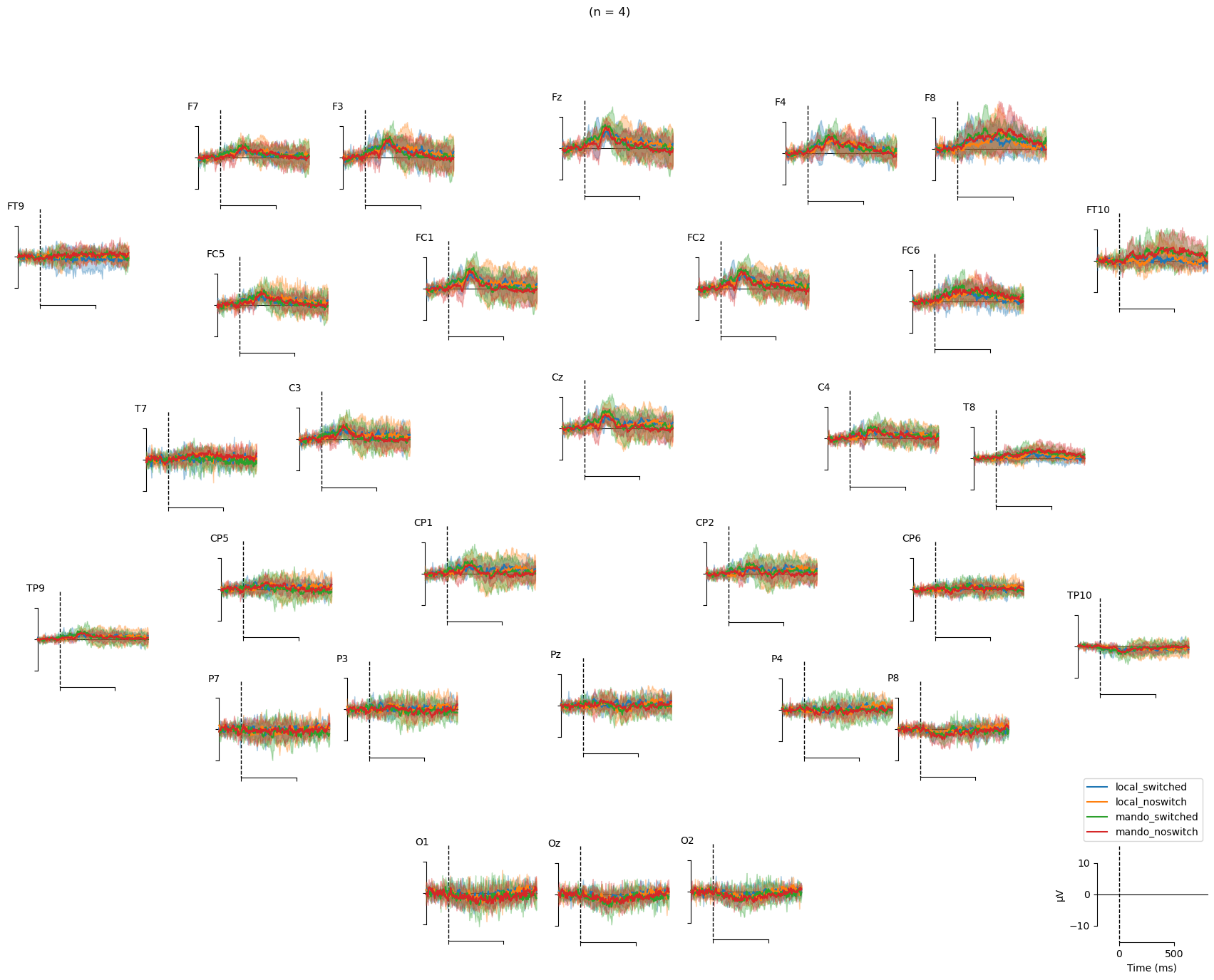
#### Topographical subplots ####
# figure title for the waveform
waveform_title = '(n = ' + str(len(sub_ppts)) + ')'
# waveforms across scalp topo
# NOTE: I don't know how to save these plots using the code
fig = mne.viz.plot_compare_evokeds(
evokeds,
axes='topo',
# picks=pick_chans,
# combine="mean",
show_sensors=True,
# colors=colors,
title = waveform_title,
# ylim=dict(eeg=[-5, 5]),
time_unit="ms",
show=False,
);
##############################